How to Use Glow on Databricks Community Edition¶
Try Glow on Databricks for free with Databricks Community Edition.
Step 1: Sign up for Community Edition¶
Sign up for a free Databricks trial with Databricks Community Edition.
Fill out the Databricks free trial form and click “Sign Up”.
In the “Community Edition” section, click “Get Started”.
Click the “Get Started” link in your “Welcome to Databricks” verification email.
Set your password and click “Reset password”. You will be redirected to your Databricks Community Edition workspace.
Step 2: Enable the Databricks Runtime for Genomics¶
Enable the Databricks Runtime for Genomics in your Databricks Community Edition workspace. The Databricks Runtime for Genomics includes Glow.
Log into your Databricks Community Edition workspace.
Click the user account icon in the top right corner of your workspace.
Select “Admin Console”.
In the admin console, select the “Advanced” tab.
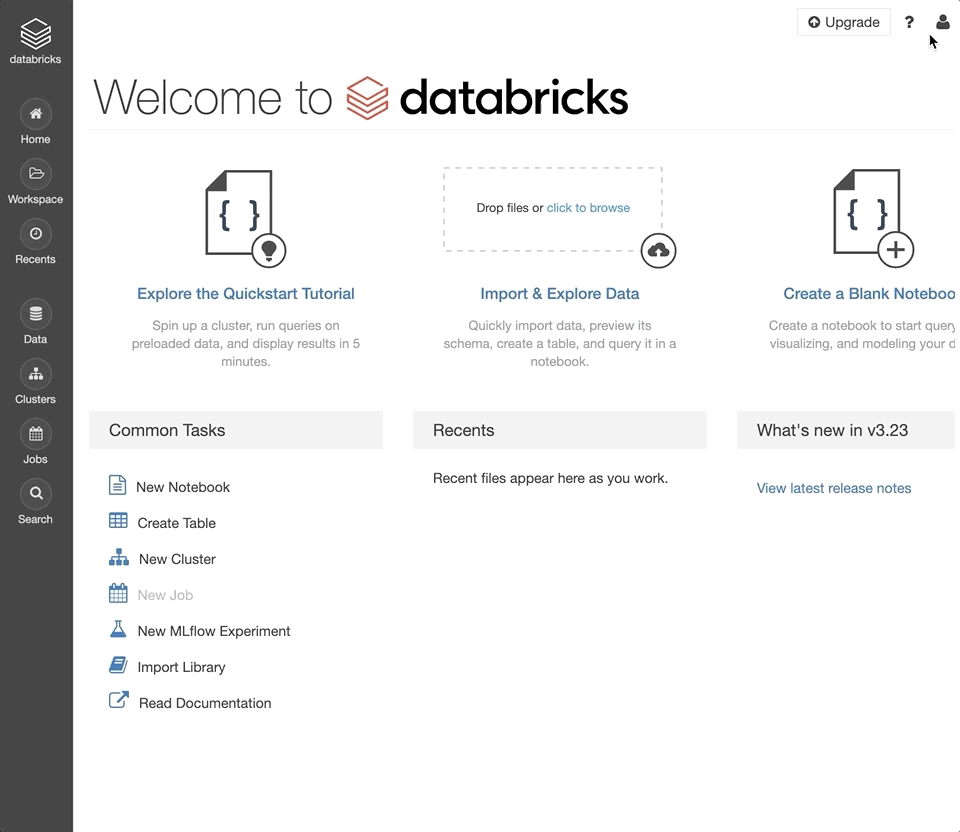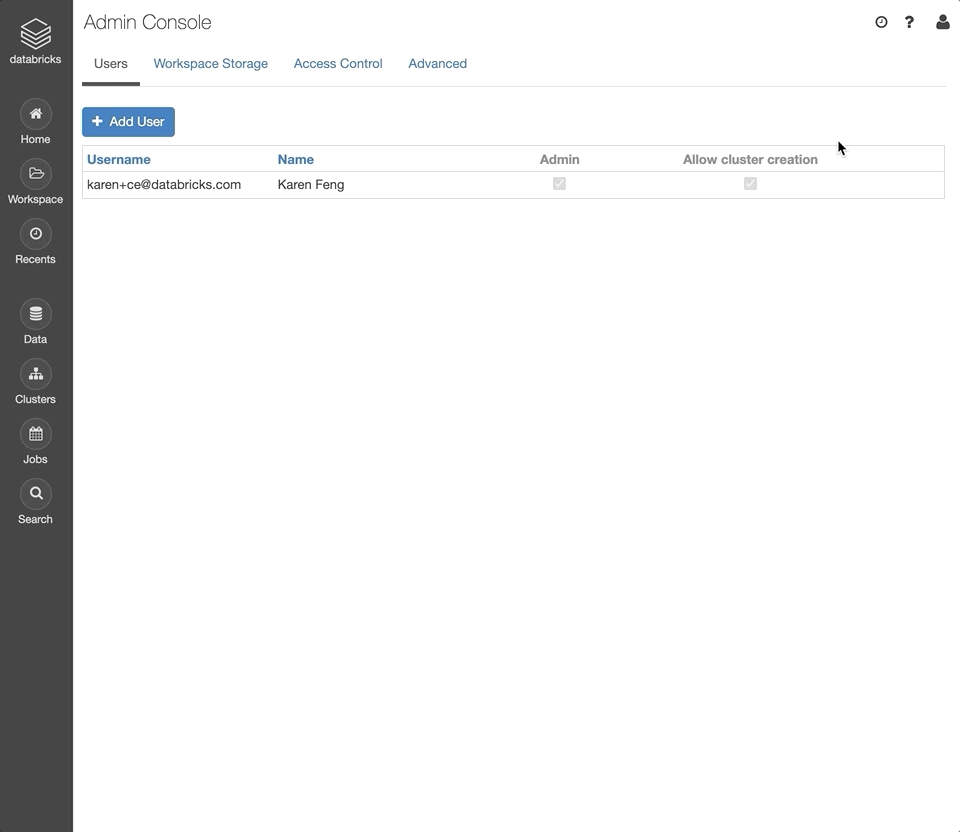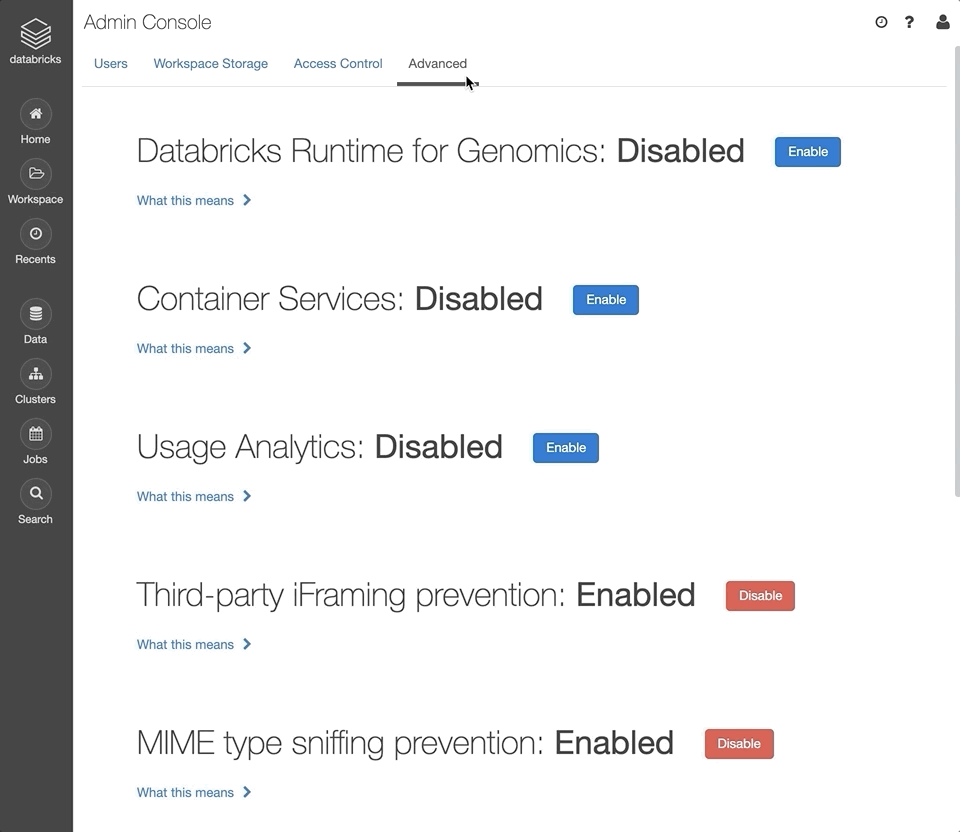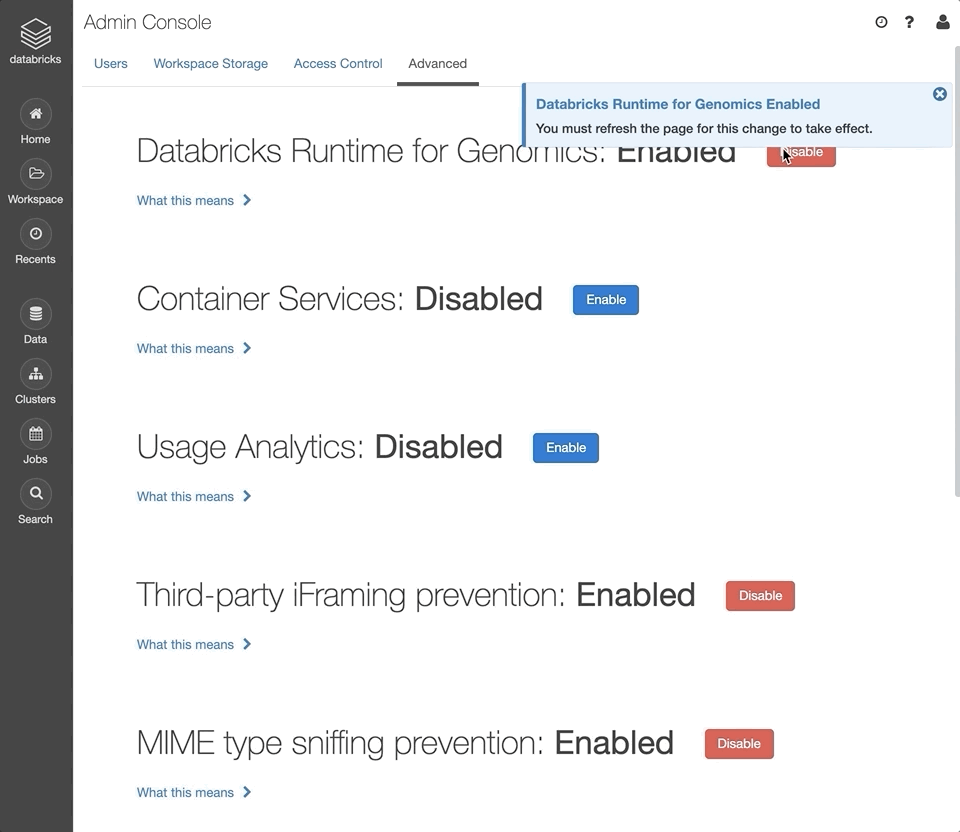
Next to the “Databricks Runtime for Genomics” section, click “Enable”.
Refresh the page for this setting to take effect.
Step 3: Import Glow notebooks¶
Import the Glow demonstration notebooks to your Databricks Community Edition workspace.
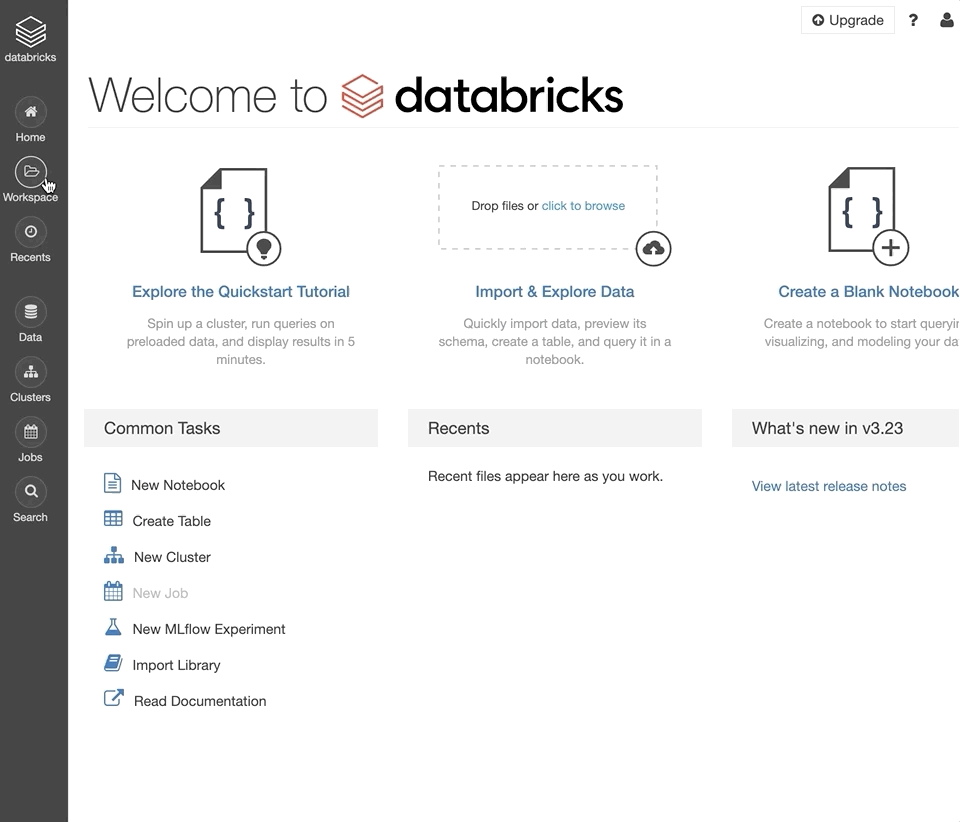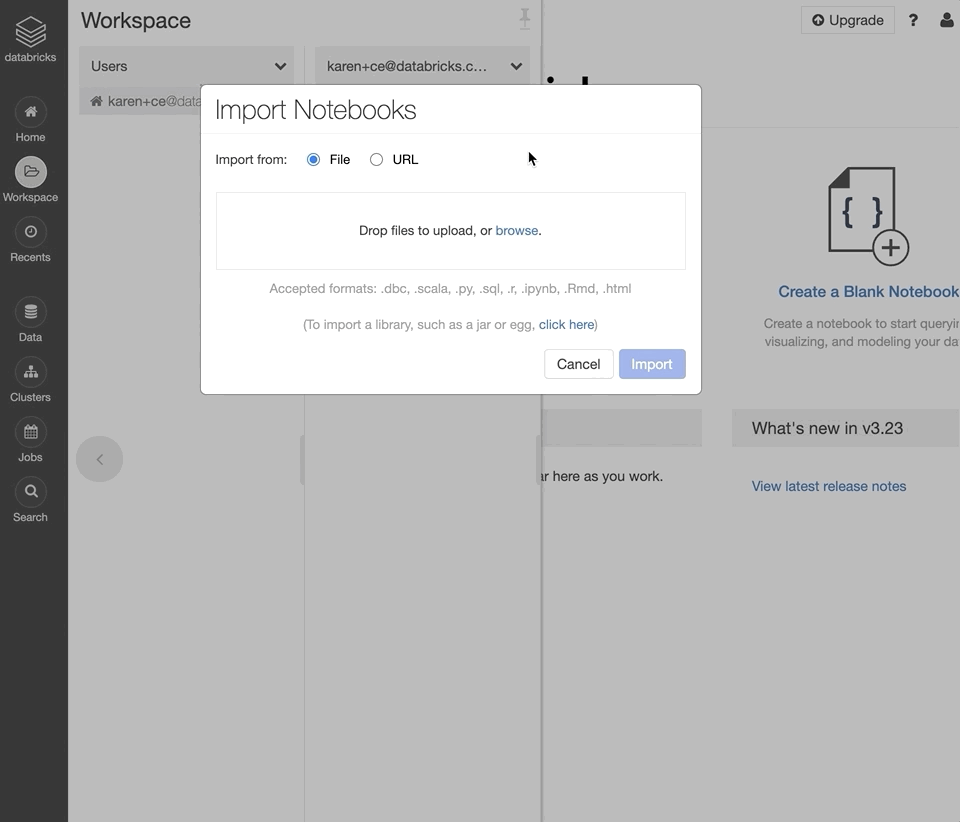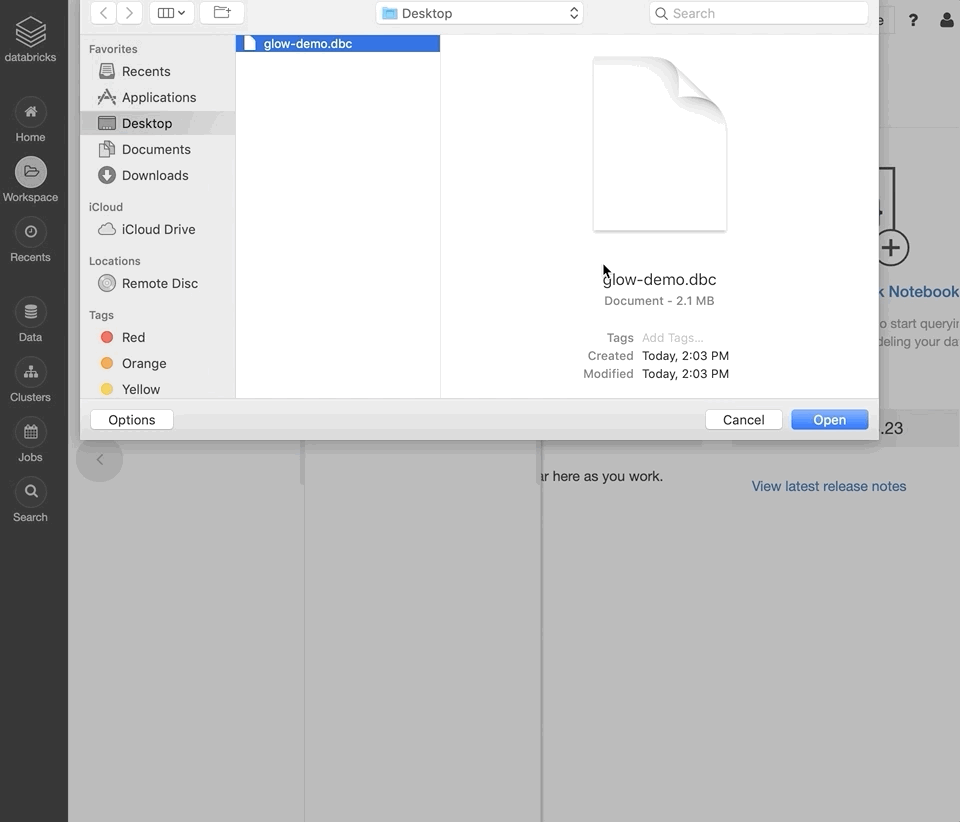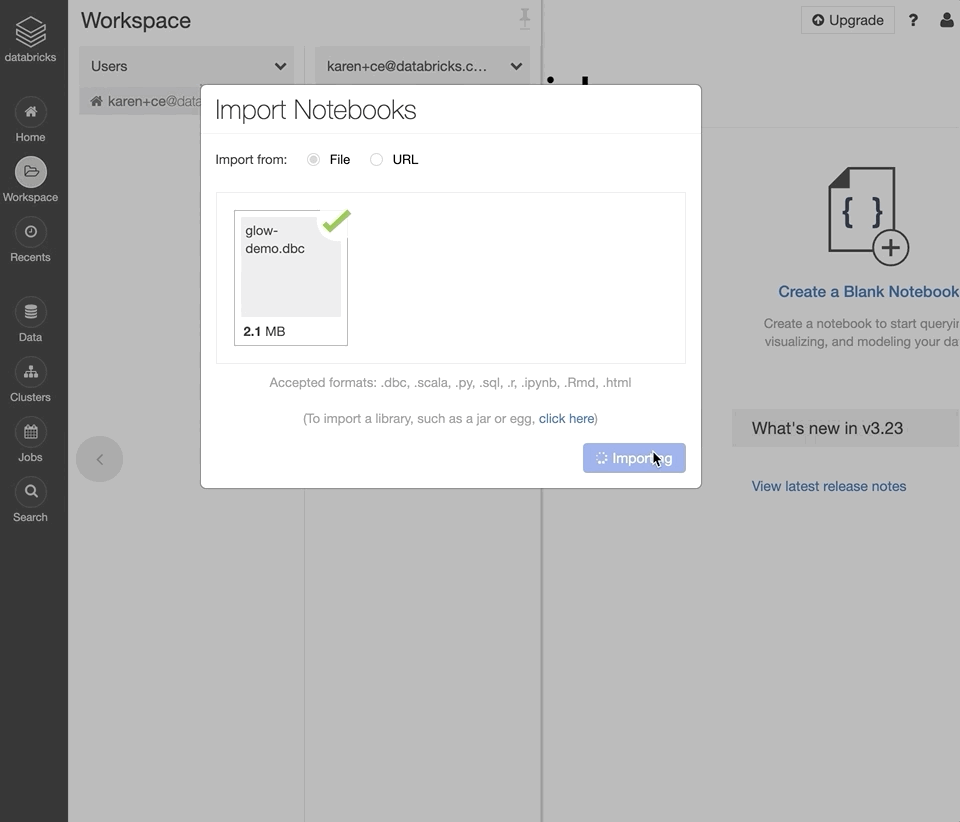
Log into your Databricks Community Edition workspace.
Download the
DBC archive.Click the Workspace button in the left sidebar of your workspace.
In your user folder, right-click and select “Import”.
Select “Import from file”, select the downloaded DBC archive, and click “Import”.
Step 4: Create a cluster¶
Create the cluster shortly before you run the notebooks; the cluster will be automatically terminated after an idle period of 2 hours.
Log into your Databricks Community Edition workspace.
Click the Clusters button in the left sidebar of your workspace.
Set the “Cluster Name” as desired.
Under the “Databricks Runtime Version” dropdown, select the latest version of the Genomics Runtime.
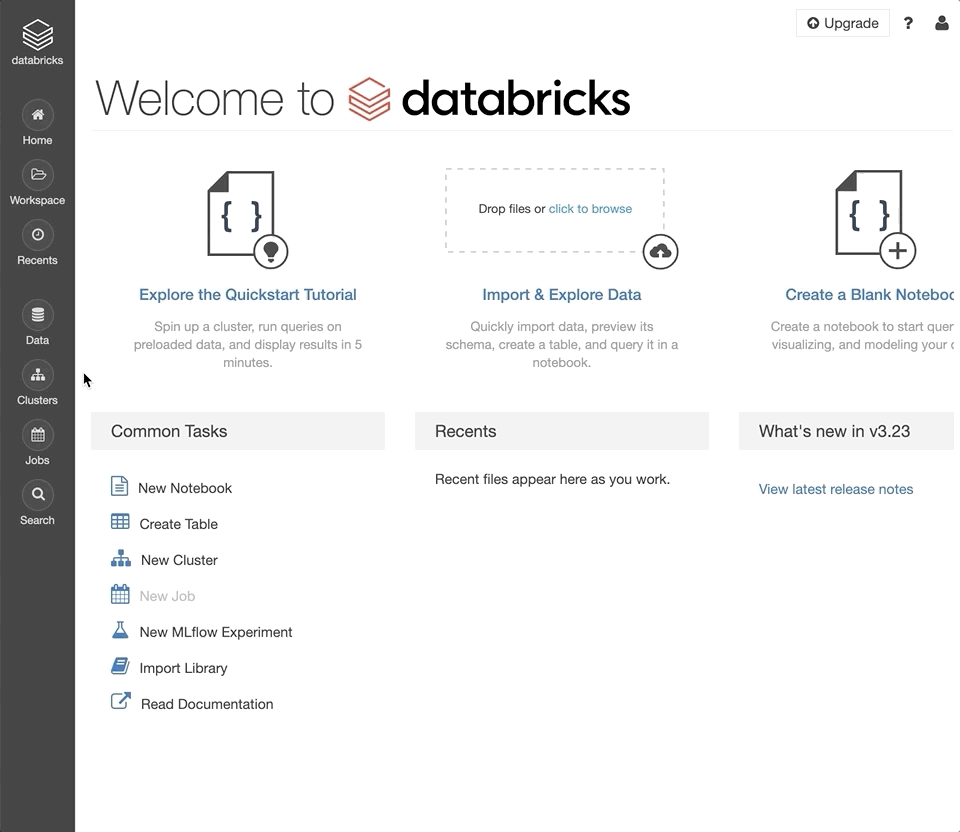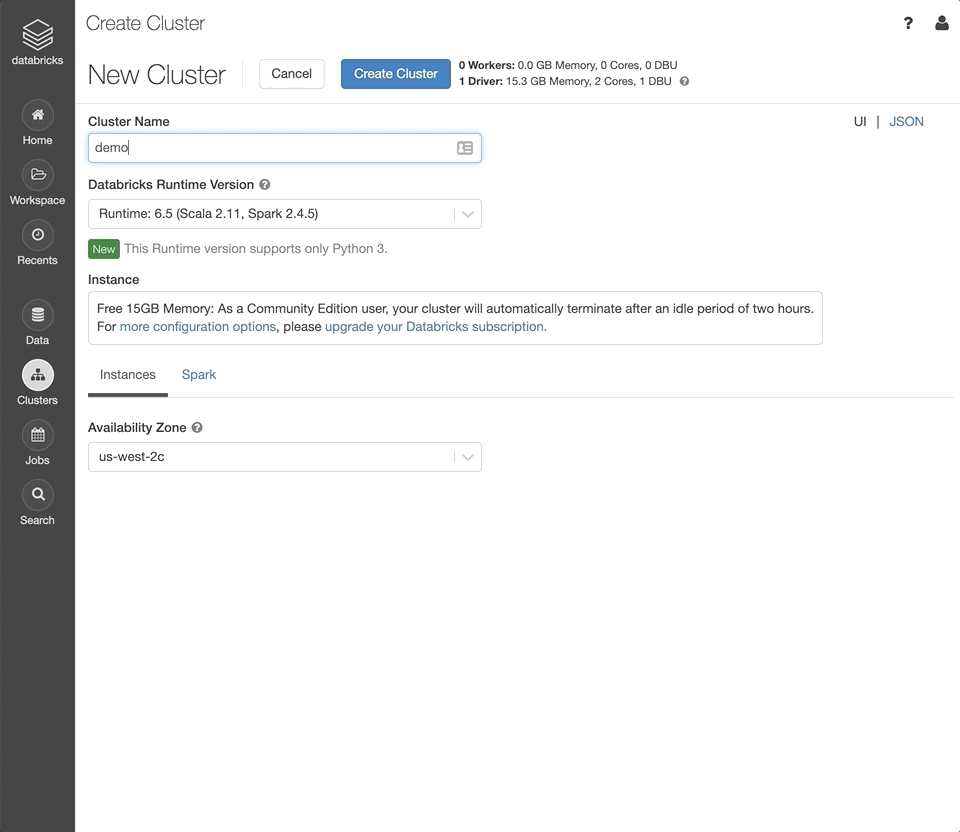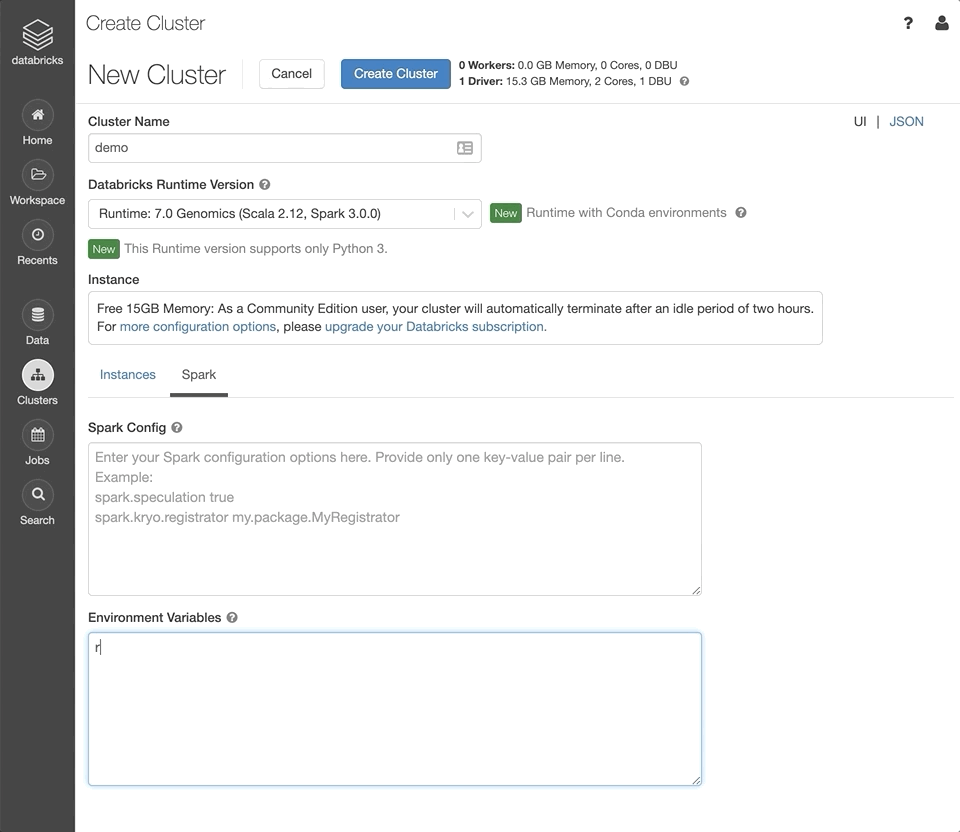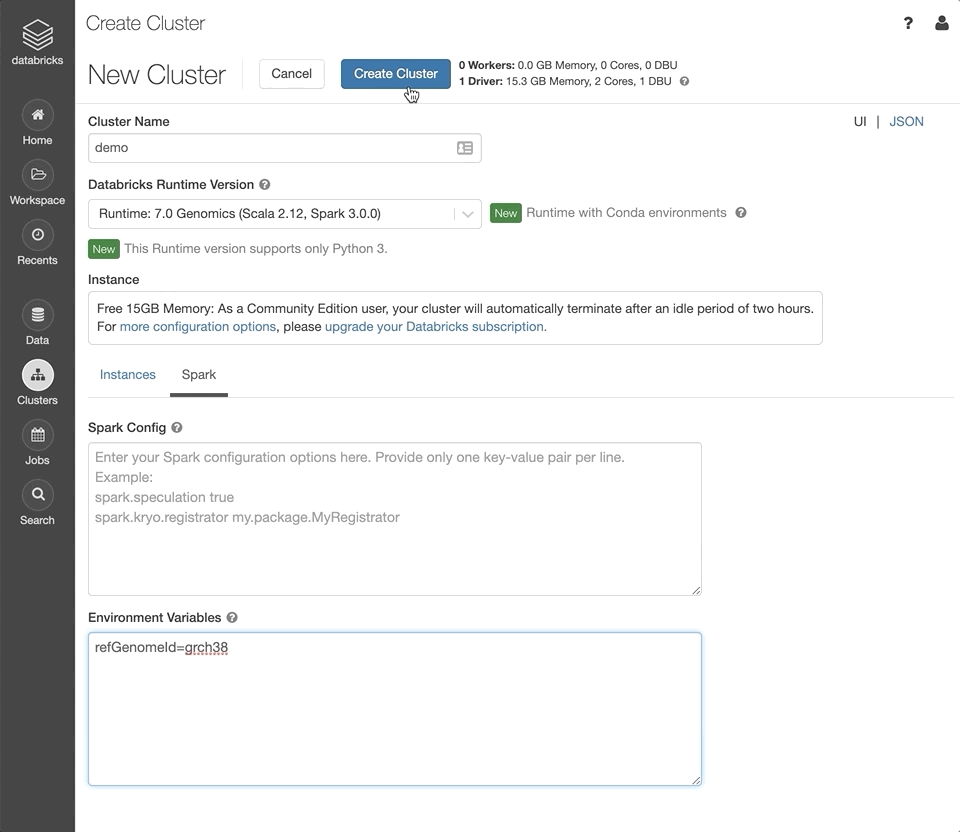
Under the “Spark” tab, set the Environment Variable
refGenomeId=grch38. This will initialize the cluster with human genome assembly 38 installed.Click “Create Cluster”.
Refresh the page to see your new cluster in the list.
Step 5: Attach cluster-scoped libraries¶
Install libraries to a cluster in order to run third-party code.
Log into your Databricks Community Edition workspace.
Click the Clusters button in the left sidebar of your workspace.
Click an existing cluster.
Select the “Libraries” tab.
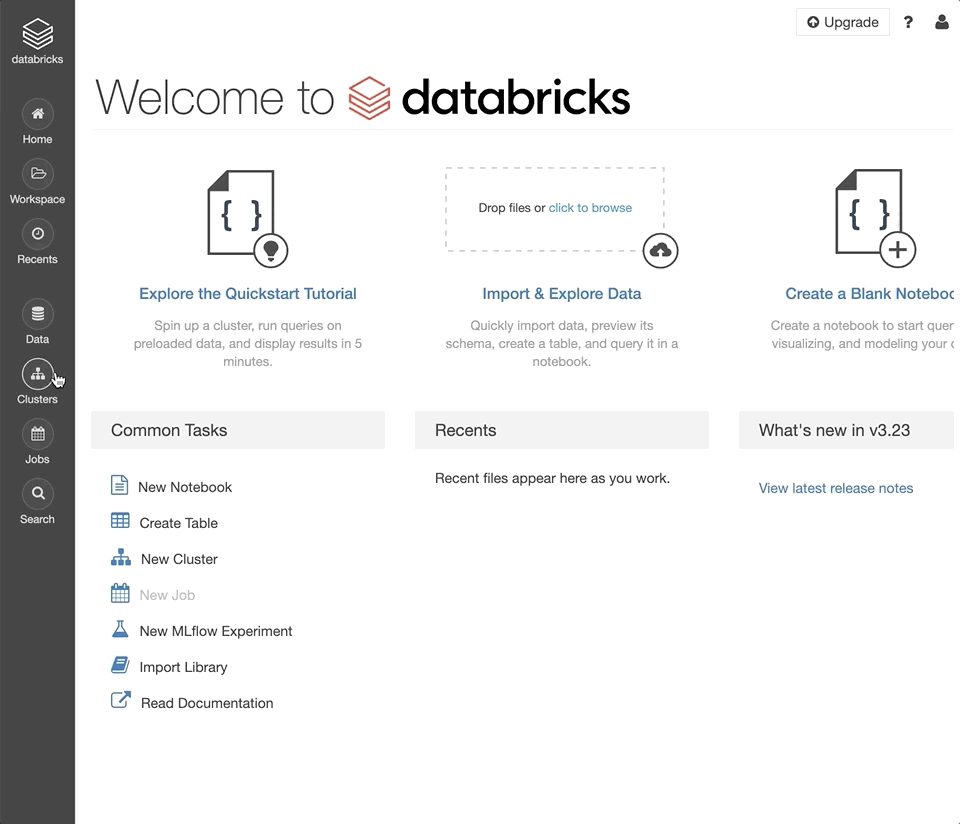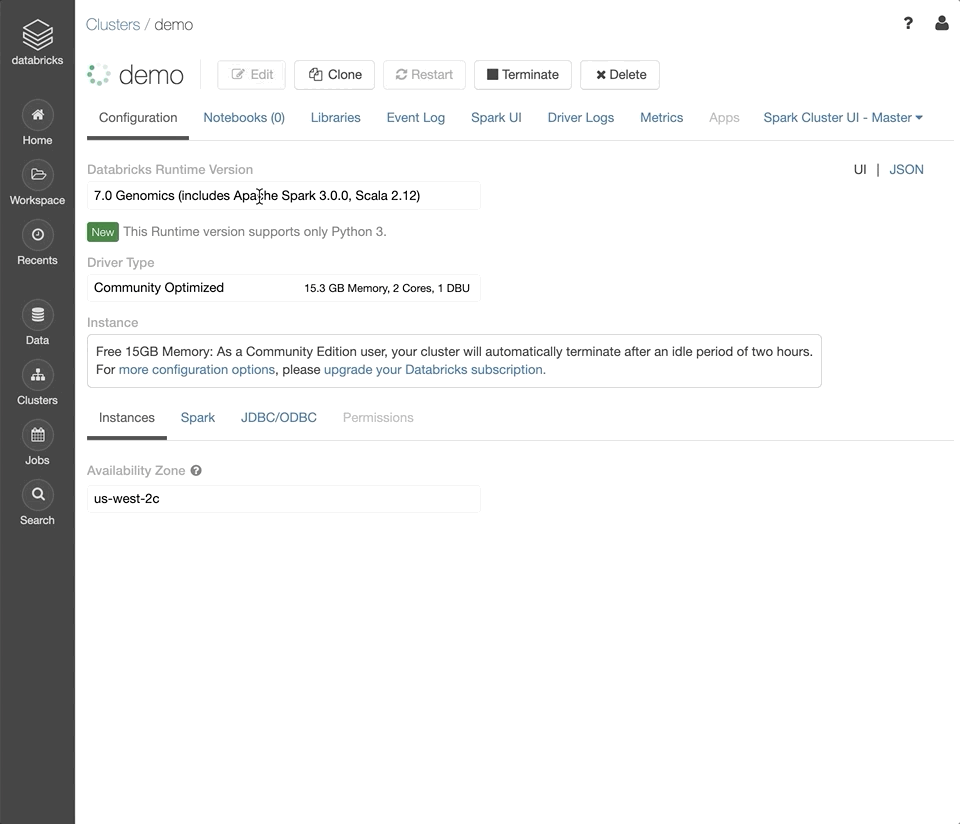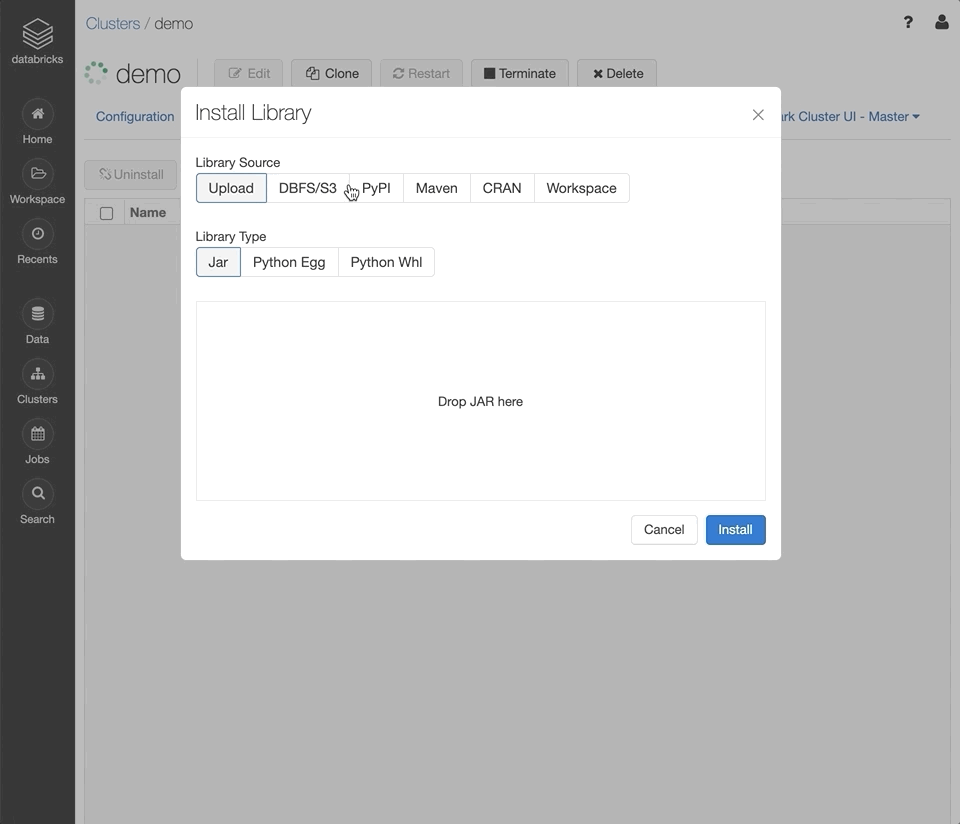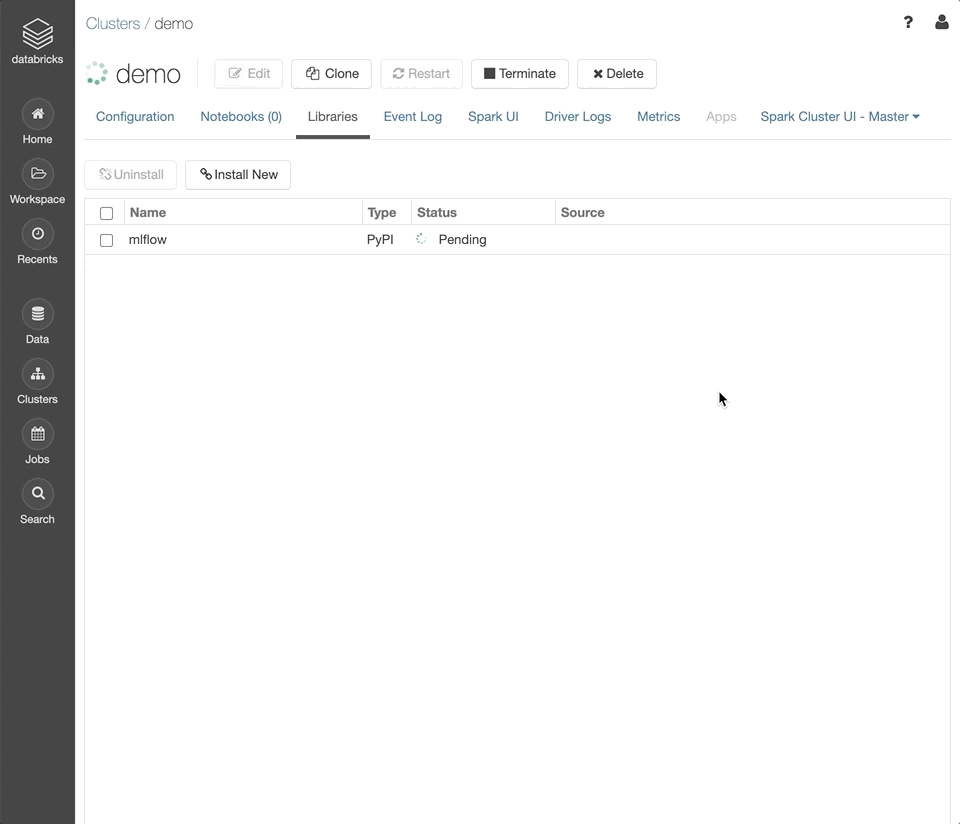
Click “Install New.”
Set the “Library Source” to “PyPi” and set the “Package” field to
mlflow. Click “Install”.Repeat the previous steps with the library
bioinfokit.